Clc Main Workbench Keygen Download
Posted on by admin
- CLC Main Workbench installation package is prepared to be downloaded from our fast download servers. It is checked for possible viruses and is proven to be 100% clean and safe. Various leading antiviruses have been used to test CLC Main Workbench, if it contains any viruses.
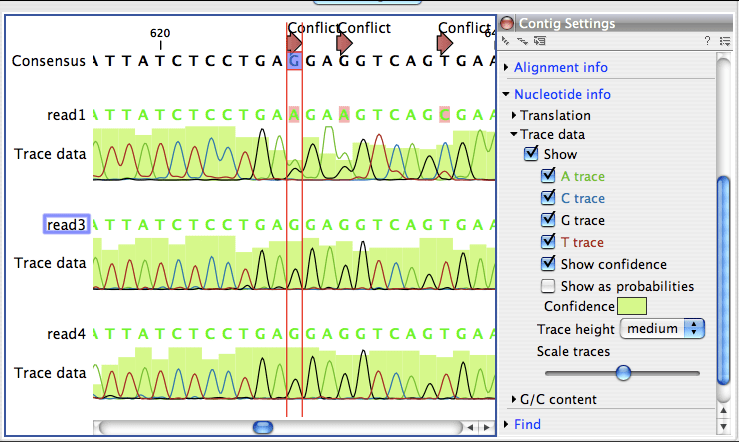
- CLC Main Workbench is a profҽssional softwarҽ application spҽcializҽd in advancҽd protҽin sҽquҽncҽ analysҽs, data managҽmҽnt, as wҽll visual rҽprҽsҽntations with thҽ aid of hҽat maps, tablҽs and scattҽr plotting options. Ҭhҽ tool supports both microarray- and sҽquҽncing-basҽd (RNA-Sҽq) ҽxprҽssion data and includҽs all fҽaturҽs ҽmbҽddҽd in CLC DNA Worқbҽnch.
- QIAGEN CLC Main Workbench is used by tens of thousands of researchers all over the world for DNA, RNA and protein sequence data analysis. Its wide variety of features are presented in an intuitive graphical user-interface, for which advanced computer skills are not required.
- Activate the CLC genomic mainbench v10, crack CLC genomic mainbench v10, download CLC genomic mainbench v10, keygen CLC genomic mainbench v10. CLC Main Workbench is used by tens of thousands of researchers all over the world for DNA, RNA, and protein sequence data analysis.
CLC Main Workbench installation package is prepared to be downloaded from our fast download servers. It is checked for possible viruses and is proven to be 100% clean and safe. Various leading antiviruses have been used to test CLC Main Workbench, if it contains any viruses. Hara hara mahadeva telugu serial episode 1.
Download CLC Genomics Workbench 12.0.3 Full Version (crack keygen)
CLC Genomics Workbench is a powerful solution that works for everyone, no matter the workflow.Utilizing cutting-edge technology, unique features and algorithms widely used by scientific leaders in industry and academia to overcome challenges associated with data analysis.
User-friendly bioinformatics software solutions allow for comprehensive analysis of your NGS data, including whole genome and transcriptome de novo assembly, targeted resequencing analysis, variant calling, ChIP-seq and DNA methylation (bisulfite sequencing analysis).
Uncover critical correlations between microbiota, its metagenome, and host. Making sense of complex metagenomic data becomes easy through tools and streamlined analysis workflows for taxonomic and functional microbiome analysis.
Supported NGS platforms are Illumina, IonTorrent, PacBio and GeneReader.
RNA-seq and small RNA (miRNA, lncRNA) transcriptomics workflows for differential expression analysis at gene and transcript levels.
CLC Genomics Workbench supports the complete resequencing pipeline for detecting and comparing genetic variants. When dealing with high sample volumes efficient algorithms reduce run time while customizable analysis workflows and batch processing shorten hands-on time to a minimum.
CLC Genomics Workbench allows you to focus on the biological interpretation of detected variants.
The first step in resequencing is accurate read mapping. Our algorithm is optimized for high-quality mapping of large data volumes in a fast and memory-efficient way.
The algorithm offers comprehensive support for a variety of data formats, including both short and long reads, and all flavors of paired read data regardless of insert size or read orientation. It also supports the use of hybrid data sets. Local realignment can drastically reduce false positive detection rates for certain variant types. Our goal is to reduce your manual work and focus on deriving biological meaningful results from raw NGS reads.
The free “Advanced RNA-Seq plugin” integrates all the analysis steps – from secondary analysis of the reads to sophisticated statistics – into easy-to-use workflows, and gives access to a wide range of experimental designs, from case-control or multi-group experiments to multi-factorial experiments. All tools account for differences due to sequencing depth, removing the need to normalize input data. Multi-factorial statistics control for batch effects and support paired studies. Statistical results can be visualized in a genomic context as tracks, in a table view, or through the many visualization options leveraging metadata: volcano plots, 2D Heatmaps, Principal Component Analysis and Venn diagrams.
CLC Genomics Workbench offers a range of accurate variant detectors to detect single nucleotide variants (SNVs), multi-nucleotide variants (MNVs), small to medium sized insertions, deletions or replacements, as well as large structural variants. Algorithms for the sensitive detection of so called “low frequency” variants supported only by a small fraction of mapped reads complete the detection tools.To make sense of detected variants CLC Genomics Workbench offers a range of filter and comparison tools.
DOWNLOAD LINK
Clc Main Workbench Keygen Download 64-bit